Exercise XI: Embeddings#
Learned Representations & The Geometry of Meaning#
Introduction#
An embedding is a dense representation where we specifically enforce a geometrical property: similar objects must be closer together in the latent space. This is different from standard dimensionality reduction; here, the “meaning” is learned through specific loss functions, such as contrastive learning.
A primary example of this is VGGish, which converts audio spectrograms into feature embeddings that a classifier can use to distinguish between complex signals, such as different types of breathing patterns.
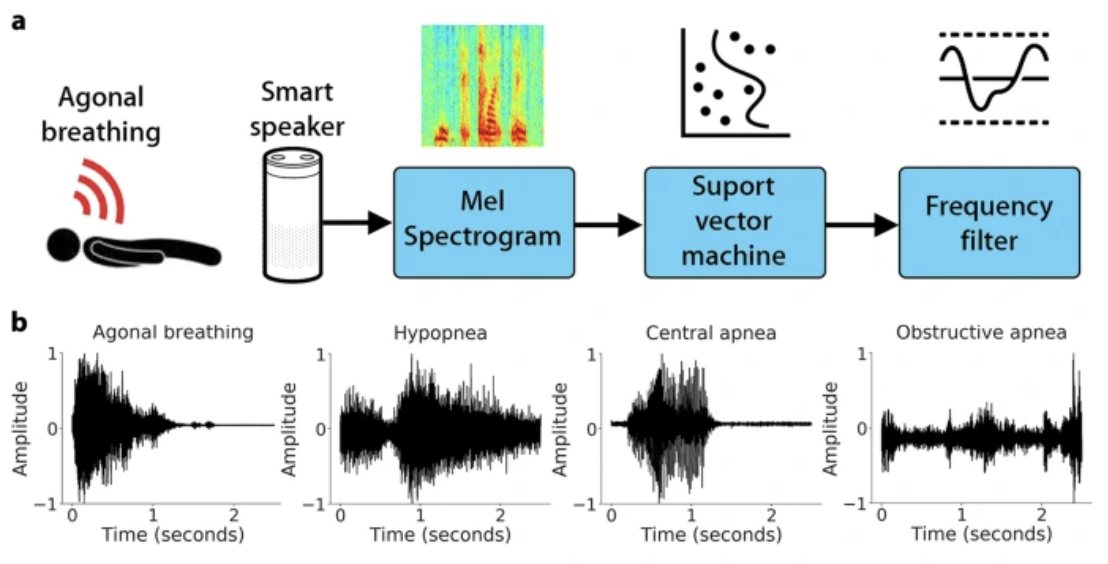
Key Concepts#
Semantic Embedding: Learning a dense representation where similar words have similar encodings.
Encoders: Models like DistilBERT (text), VGGish (audio), or CLIP (image/text) act as the “engine” that creates these vectors.
Contrastive Alignment: Models like CLIP align different modalities (images and text) into the same embedding space.
Working with Text Embeddings (DistilBERT)#
introduction to DistilBERT#
DistilBERT is a pre-trained transformer model that maps text into a 768-dimensional latent space. Each sentence is tokenized and passed through the model to produce a vector that captures its semantic essence.
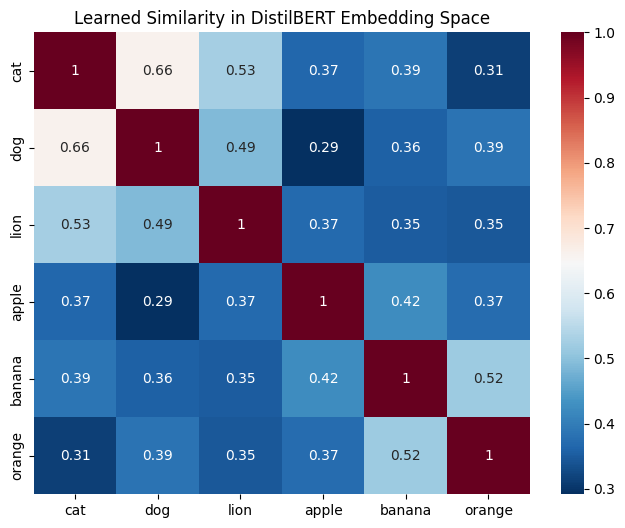
We can verify the “enforced geometry” of the embedding space by calculating the Cosine Similarity between different concepts.
from sentence_transformers import SentenceTransformer
from sklearn.metrics.pairwise import cosine_similarity
import seaborn as sns
import matplotlib.pyplot as plt
# Load a pre-trained DistilBERT-based model
# This model has already 'learned' the semantic structure of language
model = SentenceTransformer('all-MiniLM-L6-v2')
# Define stimuli categories
stimuli = ["cat", "dog", "lion", "apple", "banana", "orange"]
# 1. Generate Embeddings (Model Input -> Encoding -> Dense Vector)
embeddings = model.encode(stimuli)
# 2. Compute Similarity Matrix
# This reveals how the model clusters 'Animals' vs 'Fruits'
sim_matrix = cosine_similarity(embeddings)
# 3. Visualize the geometry
plt.figure(figsize=(8, 6))
sns.heatmap(sim_matrix, xticklabels=stimuli, yticklabels=stimuli, annot=True, cmap='RdBu_r')
plt.title("Learned Similarity in DistilBERT Embedding Space")
plt.show()
Supervised Decoding - Predicting from Embeddings#
Introduction#
Once we have these learned features, we can use them as inputs for supervised models (e.g., Logistic Regression). In this section, we predict the sentiment of a sentence based solely on its position in the embedding space.
Task: Sentiment Analysis on the SST-2 Dataset#
We will use samples from an open-source sentiment dataset to train a classifier.
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
import pandas as pd
splits = {'train': 'data/train-00000-of-00001.parquet', 'validation': 'data/validation-00000-of-00001.parquet', 'test': 'data/test-00000-of-00001.parquet'}
df = pd.read_parquet("hf://datasets/stanfordnlp/sst2/" + splits["train"])
# sample 3000 rows for demo purposes
df = df.sample(n=3000, random_state=42).reset_index(drop=True)
# 1. Prepare a small labeled dataset for sentiment analysis
sentences = df['sentence'].tolist()
y = df['label'].tolist() # 0 = negative, 1 = positive
# demonstrate "positive" and "negative" examples
rand_pos = df[df['label'] == 1]['sentence'].sample(3).values
rand_neg = df[df['label'] == 0]['sentence'].sample(3).values
print("Positive Examples:")
for sent in rand_pos:
print(f" - {sent}")
print("\nNegative Examples:")
for sent in rand_neg:
print(f" - {sent}")
Positive Examples:
- a remarkable and novel
- have been kind enough to share it
- real narrative logic
Negative Examples:
- supermarket tabloids
- lee 's character did n't just go to a bank manager and save everyone the misery
- a choppy ending
# 2. Generate Embeddings (768 dimensions per sentence)
print("Generating embeddings for 3000 SST-2 samples...")
transformer = SentenceTransformer('all-MiniLM-L6-v2')
X_embeddings = transformer.encode(sentences)
print(f"Feature Matrix Shape: {X_embeddings.shape} (Samples, Embedding Dim)")
Generating embeddings for 3000 SST-2 samples...
Feature Matrix Shape: (3000, 384) (Samples, Embedding Dim)
# 3. Split into Training and Testing sets (75% / 25%)
from sklearn.metrics import ConfusionMatrixDisplay
from sklearn.metrics import RocCurveDisplay
X_train, X_test, y_train, y_test = train_test_split(
X_embeddings, y, test_size=0.25, stratify=y, random_state=42
)
# 4. Train a Logistic Regression Classifier
clf = LogisticRegression(max_iter=1000)
clf.fit(X_train, y_train)
# 5. Evaluate the results
y_pred = clf.predict(X_test)
# 6. Visualize the Confusion Matrix and ROC Curve
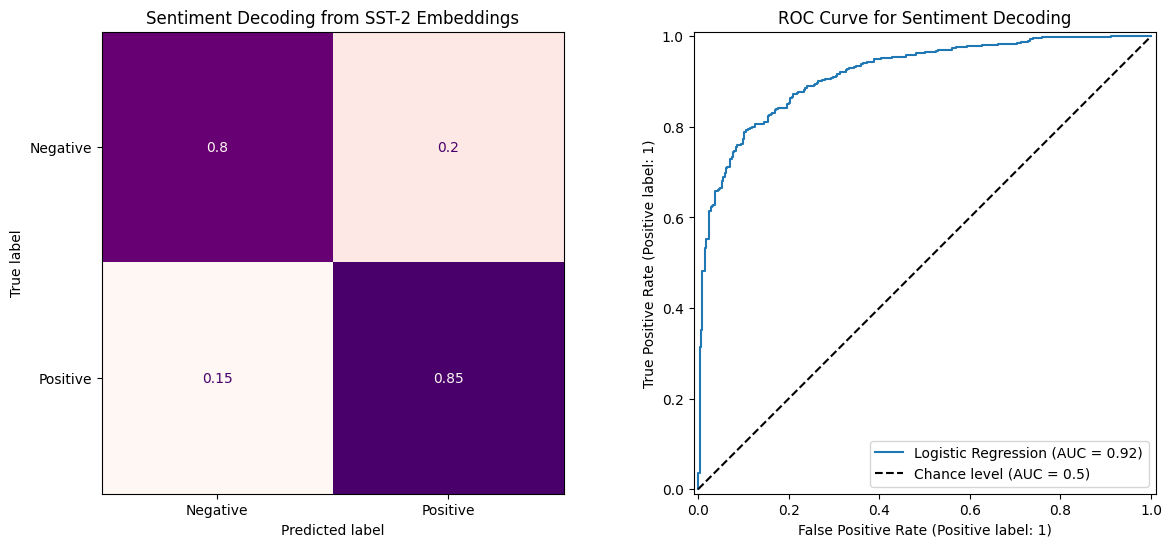
fig, axes = plt.subplots(1, 2, figsize=(14, 6))
ConfusionMatrixDisplay.from_estimator(clf, X_test, y_test,
display_labels=['Negative', 'Positive'],
cmap='RdPu',
normalize='true',
ax=axes[0],
colorbar=False
)
axes[0].set_title("Sentiment Decoding from SST-2 Embeddings")
axes[0].grid(False)
RocCurveDisplay.from_estimator(clf, X_test, y_test, ax=axes[1], name='Logistic Regression', plot_chance_level=True)
axes[1].set_title("ROC Curve for Sentiment Decoding")
Text(0.5, 1.0, 'ROC Curve for Sentiment Decoding')
From Mathematical Embeddings to Neural Representations (RSA)#
Embeddings as a Simulation of Semantic Similarity#
In this course, we treat embeddings as a mathematical approach trying to simulate the “similarity” in the way the brain processes objects of similar context or semantics. While raw brain activity (e.g., fMRI or ECoG) is a biological response, machine learning models use specific loss functions to enforce a geometrical structure where meaningful relationships are preserved in the distance between vectors.
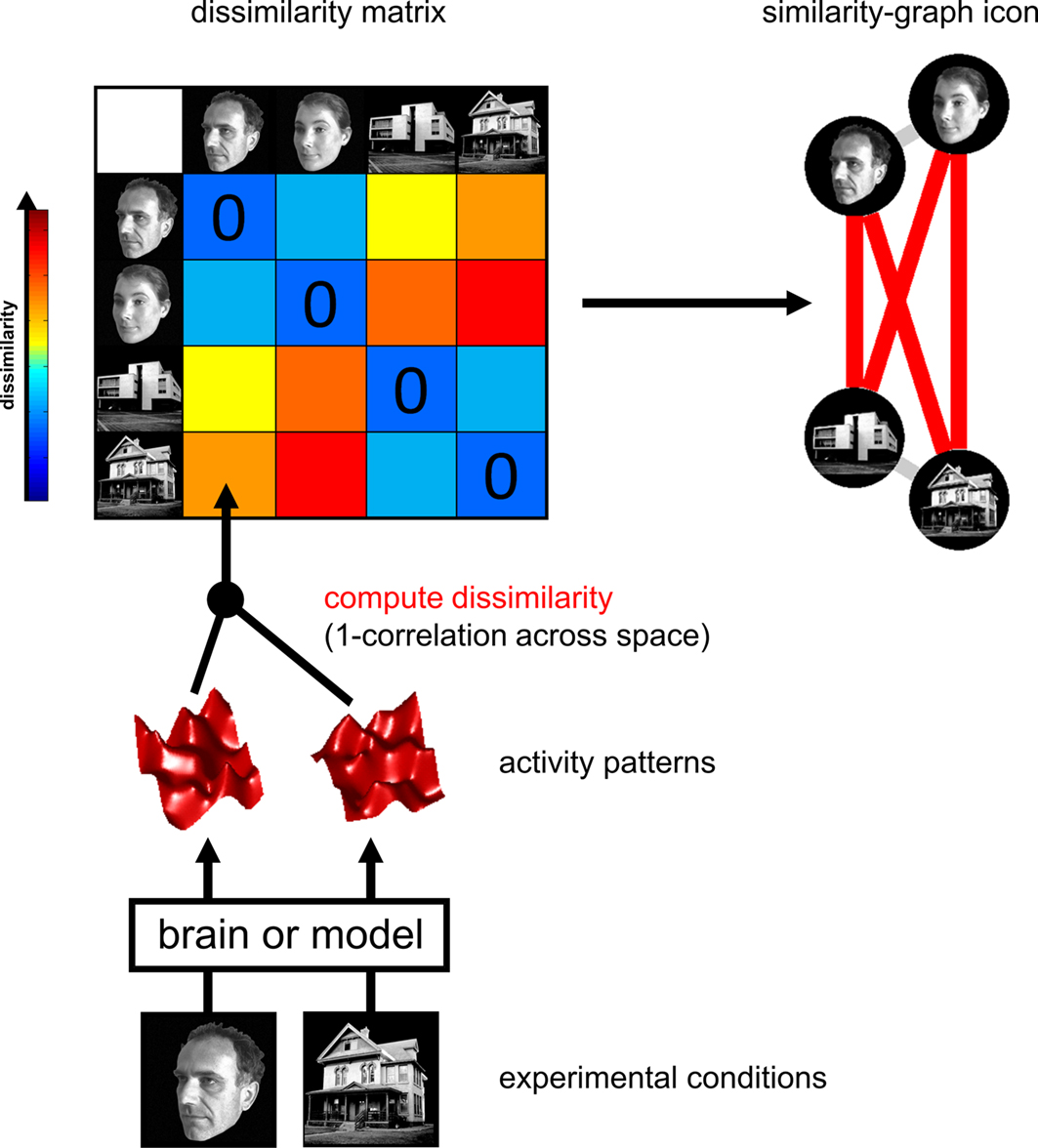
Representational Similarity Analysis (RSA):
To test if these mathematical simulations match the brain, we use Representational Similarity Analysis (RSA)\(^1\).
Model Similarity: We create a similarity matrix (like in Part 2) showing how the model represents a set of stimuli.
Brain Similarity: We create a second similarity matrix based on neural responses to those same stimuli.
Comparison: We correlate these two matrices. A high correlation suggests that the machine embedding captures a “brain-like” representation of the information.
⚠️ A Final Distinction#
Neural Activity: This is the raw biological signal. We do not automatically call it an “embedding” unless we have demonstrated that it satisfies learned geometrical properties.
Machine Embeddings: These are engineered features. We use them as a proxy or a hypothesis of how the brain might be organizing information.
[1] Kriegeskorte, N., Mur, M., & Bandettini, P. A. (2008). Representational similarity analysis-connecting the branches of systems neuroscience. Frontiers in systems neuroscience, 2, 249.
